More Information
Submitted: May 28, 2022 | Approved: June 07, 2022 | Published: June 08, 2022
How to cite this article: Bekele T, Alamnie G. Treatment of antibiotic-resistant bacteria by nanoparticles: Current approaches and prospects. Ann Adv Chem. 2022; 6: 001-009.
DOI: 10.29328/journal.aac.1001025
Copyright License: © 2022 Bekele T, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Antibiotic resistance; Horizontal gene transfer; Nanoencapsulation; Nanoparticles
Abbreviations: AgNPs: Silver Nanoparticles; AuNPs: Gold Nanoparticles; ARB: Antibiotic-Resistant Bacteria; ARGs: Antibiotic Resistance Genes; CuONPs: Copper Oxide Nanoparticles; HGT: Horizontal Gene Transfer; MDR: Multi-Drug Resistance; MRSA: Methicillin-Resistant Staphylococcus Aureus; NPs: Nanoparticles; ROS: Reactive Oxygen Species; CTiO2NPs: Titanium Dioxide Nanoparticles; VRSA: Vancomycin Resistance Staphylococcus Aureus; ZnONPs: Zinc Oxide Nanoparticles
Treatment of antibiotic-resistant bacteria by nanoparticles: Current approaches and prospects
Tigabu Bekele1* and Getachew Alamnie2,3
1Department of Chemistry, Mekdela Amba University, Tuluawuliya, Ethiopia
2Department of Biology, Mekdela Amba University, Tuluawuliya, Ethiopia
3Department of Biology, Ph.D. Candidate, University of Gondar, Industrial Biotechnology, Ethiopia
*Address for Correspondence: Tigabu Bekele, Department of Chemistry, Mekdela Amba University, Tuluawuliya, Ethiopia, Email: [email protected]
Antibiotic-resistant bacteria are emerging pathogens whose resistance profiles generate a serious health crisis by holding their impact on human health. Misuse of antibiotics has directed the emergence of microbes immune to presently accessible drugs. Pathogenic bacteria become resistant by employing various mechanisms, such as; antibiotic modification, target site alteration, and biofilm formation, increasing the time they spend in the intracellular environment where antibiotics are unable to succeed at therapeutic levels. Due to this, attempts are being made to develop new alternative nanoantibiotics as a promising approach to treat multidrug resistance disease-causing bacteria. Accordingly, there is considerable contemporary attention to the use of nanoparticles (NPs) as antibacterial agents against different pathogens and as target drug delivery toward specific tissues therefore microbes are eliminated by the biocidal properties of nanoantibiotics. Additionally, the utilization of nanoencapsulation systems can help to beat the issues of, those with toxicity natures, and target drug delivery problems. This review encompasses the antibiotic resistance prevalence, mechanisms, and therefore the use of nanoparticles as antibacterial and drug delivery systems to overcome the antibiotic resistance challenges of bacteria. Overall, this review paper provides a conceptual framework for understanding the complexity of the matter of emergence of antibiotic resistance bacteria even for brand spanking new synthesized antibiotics. Therefore the availability of such knowledge will allow researchers to supply detailed studies about the applications of nanoparticles in the treatment of multidrug-resistant bacteria.
Multi-drug resistance is a serious problem to global community well-being and has attracted worldwide attention [1,2]. Antibacterial resistance by bacterial pathogens becomes increasing because of mutations after exposure to various drugs leading to infections in healthcare centers and communities [1,3-5].
Large numbers of pathogens are getting multidrug-resistant (MDR) due to inadequate dosage, long-term usage, evolvability of microbes, and intensive and inappropriate application of antibiotics in veterinary, clinical, and agricultural settings, leading to the detection of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) in diverse settings [4,6-10]. This is when patients stop their medication dose by taking half in concentration or number (capsule) on their own self and the amount taken may not kill all the number of pathogens satisfactorily because of low/inadequate dosage of drugs. Through this, pathogens become adapted to the prescribed drugs and became active after a time and do it best of all by adapting to the medication. Thus, evolutionary biology suggests drug resistance is a factor in the emergence and spread of drug resistance. This problem may be a patient’s error. Health risk expectations of drug resistance are challenging as there are continuous additions of the latest antibiotics from different environments, that obscure the antibiotic resistance mechanisms encoded by its resistome [2,3,5].
Recent reports have indicated that horizontal gene transfer (HGT) has become a major concern in public health through the spread of antibiotic resistance genes amongst pathogenic bacteria [1,2,10]. Mobile genetic elements like transposons and plasmids can disseminate a various range of antibiotic resistance genes [11]. Bacteria developed resistance through diverse mechanisms, for example, drug target alteration, degradation of antibiotics, and reduction of antibiotic uptake [12]. The currently available antibiotics may be found in low concentrations at the injection site and need multiple entry mechanisms to sustain an endless bactericidal effect [10]. To mitigate this, recent efforts in addressing the antibiotic resistance challenges lie within the use of nanoparticles as antimicrobial agents against different pathogens that resist multidrug and as antimicrobial delivery vectors toward specific tissues [9].
Nanoantibiotics and nano delivery systems are a relatively new but rapidly developing science where materials within the nanoscale range are employed to serve as means of antibacterial and deliver therapeutic agents to specifically targeted sites in a controlled manner [13,14]. Nanoparticles have an antibacterial nature by overcoming existing drug resistance mechanisms like destructing biofilm formation and inhibiting biomolecule synthesis [13,15].
Nowadays, the conventional antimicrobial delivery system causes microbes to develop different resistance mechanisms, and one of the foremost encouraging approaches to advance the efficacy of antibiotics is to compound them with nano delivery materials [13,16]. Such vectors can protect drugs from enzyme degradation and increase the satisfying efficacy of the drugs [17]. Therefore, the objective of this review is to report on new insights about employing nanotechnology, specifically nanoantibiotics and nano-drug delivery as a fresh paradigm for overwhelming antibacterial resistance.
Antibiotic resistance prevalence of bacteria
The spread of ARB and ARGs became an evolving problem for worldwide health [3,5,8,13,14,18,19]. Recent reports have discussed that horizontal gene transfer leads to the gaining or alteration of characters for drug-fighting and virulence factors [1,19,20]. Similarly, reported that introduced genes in one pathogenic bacterium may confer a novel trait in other bacteria, which might be a cause of possible harm to the health of individuals or the environment [12,19]. Table 1 shows antibacterial development and subsequent resistance timelines.
| Table 1: History of antimicrobial development and later development of resistance. | |
| Antibiotics | History |
| Penicillin (1942) | Penicillinase spread (1945) Transferable Penicillinase in Gonococcus (1976) Penicillin-resistant Enterococcus (1983) |
| Streptomycin (1947) | Streptomycin resistance (1947) |
| Tetracycline (1952) | Tetracycline resistance (1956) |
| Vancomycin (1958) | Rarely used until the mid-1980s Vancomycin-resistant Enterococcus (VRE) (1987) Vancomycin intermediate resistant S. aureus (ViSA) (1996) Vancomycin-resistant S. aureus (VRSA) (2002) |
| Methicillin (1959) | Methicillin-resistant S. aureus (MRSA) (1961) Community-acquired MRSA (1999) |
| Cephalothin (1964) | Cephalothin resistance: 1st cephalosporin (1966) |
| Gentamicin (1967) | Gentamicin resistance (1970) |
| Cefotaxime: FDA approved (1981) | Cefotaxime resistance (1983) The first outbreak of 3rd cephalosporin-resistant K. Pneumoniae (1987) |
| Imipenem, 1st Carbapenem (1984) | Carbapenem-resistant Acinetobacter baumani (1998) |
| Linezolid, first oxazolidinone: FDA approved (200) | Linezolid-resistant S. aureus and VRE (2001) |
Recent reports have revealed that the misuse of antibiotics in different environments leads to a global spread of resistance because microorganisms are experts at adaptation influenced by human activity followed by the natural genetic exchange [4,7,8,16,19,21,22]. Nguyen, et al. suggested that there is a growing body of evidence that indicates that high levels of antibiotic resistance emerge due to combinations of several mutations and, they get mobile genetic elements from other bacteria through genetic exchange mechanisms [2,10,19]. Recent reports have found that MDR Enterococcus isolates harboring both resistance and virulence genes in fork samples function as a reservoir for the potential transfer of those genes from poultry to humans through the farm to fork continuum [22].
Antibiotic resistance mechanisms in bacteria
The age of recent antibiotics started with the invention of penicillin by Fleming and since then, several antibiotics are introduced to save lots of lives (Table 1). However, resistance evolved over time in comeback to the unintended discharge of antibiotics to different environments [2,3,5,8], indicated that the occurrence and rise of antibiotic-resistant microbial populations in several environments for various classes of antibiotics are unavoidable due to the principles of evolution. Similarly [1], revealed that drug resistance could be intrinsic, mutations, or due to horizontal gene transfer. Examples of antibiotic-resistant bacteria that are multidrug-resistant bacteria usually of problem in healthcare practices are Mycobacterium tuberculosis, Escherichia coli, Klebsiella pneumonia, Enterococci spp., Pseudomonas aeruginosa, Staphylococcus aureus, Staphylococcus pneumonia, etc. Antibiotic resistance genes have emerged as environmental contaminants and have the capability to spread among bacteria and distribute from humans and animals to the natural environment [2,3].
Antibiotic modification degradation
Enzymatic inactivation enzymes like hydrolase and transferase enzymes lead to the inactivation of the antimicrobial agent’s structure of the drugs. Antibiotic modification or degradation is one of the foremost effective bacterial approaches to resist the actions of antibiotics. This strategy is by the production of certain enzymes that deactivate the antibiotics by adding specific chemical moieties to the compound, rendering the antibiotic unable to interact with its target [12]. Phosphorylation and adenylation are known to be employed by some pathogenic bacteria, primarily against aminoglycosides. In addition to this [12], reported that β-lactamases are the most common resistance mechanism used by gram-negative bacteria to β-lactam drugs.
Target modification
Microorganisms are quite surprising in the adaptation and evasion of molecules against them in the era of antibiotic resistance. From the mechanisms, changing the cellular targets of antibiotics is a common mechanism of antibiotic resistance within the bacterial world. Peterson and Kaur, 2018 revealed that modification in the cellular targets acts as a self-resistance tool in the contradiction of many groups of antibiotics [12]. Analysis of the biosynthesis clusters of β-lactam producing bacteria showed that they often contain genes for protein-binding proteins (PBPs), suggesting their role in self-resistance [23].
Decreased uptake and increased efflux of drugs
Proteins found in the plasma membrane-like transmembrane proteins and efflux pumps play a significant role in increasing antimicrobial resistance rendering drugs of no use [4]. This is how antibiotics have less chance of entry into the cytoplasm and efflux pumps avoid the entrance of therapeutic level antimicrobials within the microbial cell. Accordingly, has been shown that drug efflux pumps of the bacterial pathogens prevent the entry of drugs and increase the efflux of the antibacterial agent from the microbial cell before it reaches its target site to exert its effect [13]. Furthermore, many bacterial species have the capability to overexpress efflux pumps, deliberating them with an outstanding ability to battle the effects of many available antibiotics. Also, Verni, et al. reported that comparatively, gram-negative bacteria are more resistant than gram-positive this is due to the presence of a large periplasmic space along with an outer membrane, and the drug is sequestered by an efflux pump directly from the periplasm and extruded out even before they reach the cytoplasm [24]. In addition, Hemeg found that E. coli has the ability to extrude antibiotics through its many efflux pumps [13].
Biofilm formation
A biofilm is often defined as a matrix-enclosed accretion of single or multiple strains of bacteria that adhere to biological or non-biological surfaces and communicate by secreting chemicals [13,25]. Cao, et al. demonstrated the resistance nature of bacteria in biofilms are more resistant to drugs related to their free-floating state, as biofilms prevent antibiotic permeation [25]. The accumulation of many cells in biofilms may increase resistant mutants which will be designated under antibiotic pressure and therefore DNA in the extracellular matrix can enhance the transfer of resistance genes among the bacteria finally leading to bacterial resistance to brand-spanking available standard antibiotics [26]. Biofilms formed by several pathogenic bacteria result in the development of resistance against antimicrobial compounds [27]. Furthermore, Karami, et al. found that biofilms of Pseudomonas aeruginosa act as a barrier to diffusion by deceiving and degrading antibiotics and thus render tolerance to even high concentrations of antibiotics [28].
Nanotechnology in the fight against antibiotic-resistant bacteria
Nanotechnology-based on nanoscale materials is being more widely used in therapeutic settings, particularly as a new paradigm for infectious illnesses. Multidrug-resistant organisms (MDROs) infections are becoming more common as a cause of morbidity and mortality across the world. The number of antibiotics available to treat infections caused by MDROs is frequently restricted. These clinical issues emphasize the urgent need for new and more effective antibacterial methods. Nanotechnology in mitigating antibiotic resistance is based on the science of using nanoscale materials utilized for nanoantibiotics and nano-drug delivery systems. This new approach can be applied to the production of new antibiotics is a promising approach, since the use of nanometric size materials can result in greater contact between the compound and the bacteria with improved bioavailability, increased absorption, and the faster passage of the drug into the cell [14,29]. Agarwal, et al. found that there is a controlled release system for the targeted delivery of encapsulated or surface adsorbed drugs [30]. Nanomaterials can penetrate the cell wall of pathogenic microorganisms and interfere with important molecular pathways, formulating unique antimicrobial mechanisms [9]. In combination with standard antibiotics, NPs have found synergy antibacterial natures to conventional antibiotics and may aid in limiting the global crisis of emerging bacterial resistance.
Antibacterial nature of nanoparticles
Many factors influence the pharmacokinetics of NPs, including particle shape, particle size, particle surface charge, particle surface coating, nature of particle, protein binding, dosage, and animal species. NPs are now being seen as a viable alternative to antibiotics, and they seem to have a high potential for solving the issue of bacterial multidrug resistance [25]. Nanoparticles are nanosized molecules with a minimum of one dimension within the nanometer scale range (1-100 nm).
Their high antibacterial activity is due to their wide surface area to volume ratio, which enables the binding of a large number of ligands on the nanoparticle surface and thus complexation with receptors on the bacterial surface [30]. NPs have multifunctional pathways for attacking bacteria that are distinct from those of commonly available antibiotics, and they may be potential antimicrobials. Nanoparticles have the ability to penetrate bacteria and interact with a number of internal components. However, bacteria exposed to nonlethal concentrations of AgNPs showed an increase in their resistance due to mutations that caused the down-regulation of several genes [5].
Mode of action of nanoparticles as antibacterial
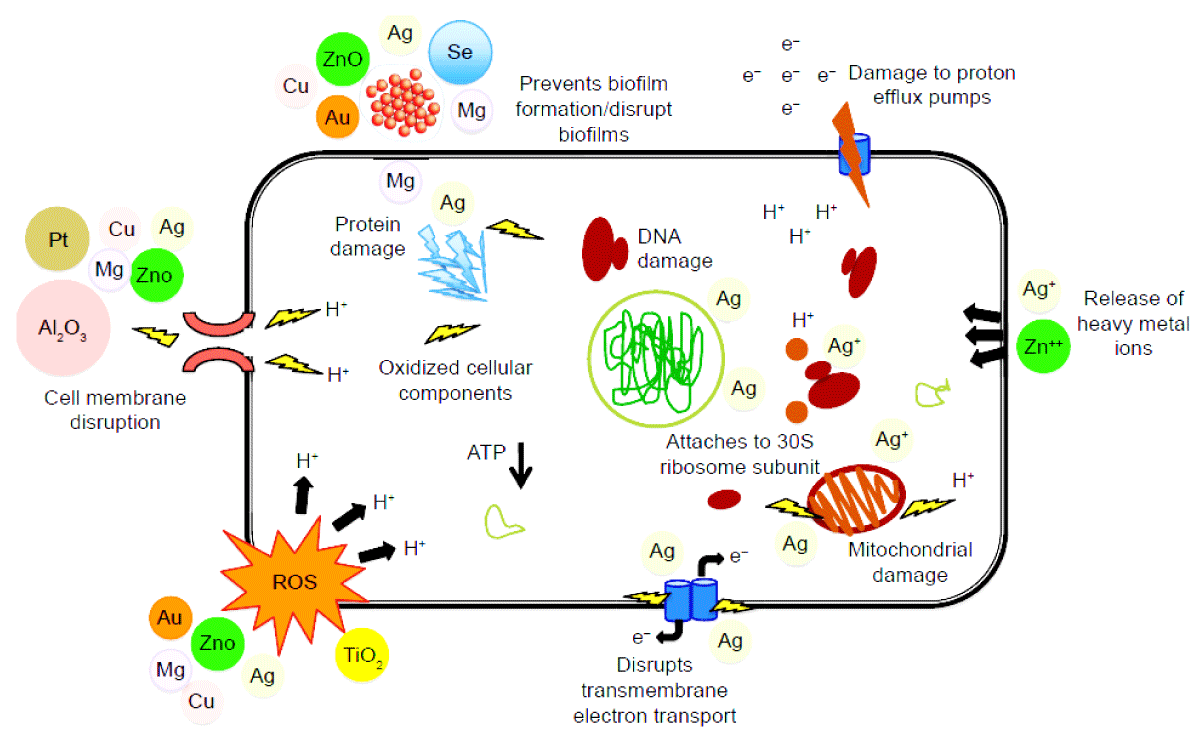
The ability of NPs to impart physical defense against bacterial resistance mechanisms improves their therapeutic potential, and they provide complicated antimicrobial mechanisms against which bacteria are unlikely to develop resistance (Figure 1). Enzymatic repression, improvements in gene expression, oxidative stress pathways, protein deactivation, and the release of free metal ions are all activated as bacteria interfere with NPs, resulting in cell wall injury, enzyme inactivation, reduced cell permeability, and disruption of DNA synthesis [13-15,31]. Also [25,28,32] revealed that NPs have the nature and characteristics to treat antibiotic-resistant biofilms.
Figure 1: Mode of action of nanoantibiotics as antibacterials adopted from [31].
Generation of reactive oxygen species (ROS)
One of the most critical mechanisms by which NPs can interrupt disease-causing agents’ normal metabolic pathways in the oxidative stress caused by ROS (Figure 1). As a result, revealed that the toxicity of NPs has been linked to the formation of reactive oxygen species (ROS) such as superoxide anions, hydroxyl radicals, and hydrogen peroxide, which inhibit DNA replication and protein synthesis while also destroying cell membranes via lipid peroxidation, compromising membrane semi-permeability, and suppressing oxidative phosphorylation [13]. As a result, the amount of ROS produced by NPs is determined by the chemical nature of the NPs. Excessive processing of ROS disrupts redox homeostasis, resulting in oxidative stress, which affects membrane lipids and changes DNA and protein structure [33] revealed that the ROS generated by nanomaterials can damage and destroy the cellular components of pathogens irreversibly (e.g., membrane, DNA, protein, and mitochondria), resulting in cell death. Moreover [19], reported that after 2 hours, intracellular ROS levels in E. coli treated with 100 mg/L Cu2O, Ag, and ZnO increased by 205 percent, 48 percent, and 136 percent, respectively.
Destruction of biofilm and its resurgence
Many studies have shown that nanomaterials can disrupt bacterial membranes and hinder biofilm production, limiting the microorganism’s ability to survive. Many methods for inhibiting biofilm formation include targeting and interfering with quorum sensing molecules. In this regard, Mahamuni-badiger, et al. found that ZnONPs destroy biofilms by releasing Zn+ ions. Figure 1 also shows the destruction of bacterial biofilm by ZnO, CuO, and AgNPs [32].
Causing direct damage
Gram-negative bacteria’s cell membrane is made up of lipid conjugated proteins and phosphates, which form a layer that only allows some macromolecules to get through. According to Anuj, et al. AgNPs have destroyed the membrane of P. aeruginosa [34]. The adhesion of nanoparticles to bacterial membranes causes the membrane to be disrupted or porosified, allowing the NPs to enter the cell and interact with essential molecules and organelles such as enzymes and DNA. The antimicrobial actions of NPs against MDROs are shown in Table 2.
| Table 2: Nanoparticles against MDR pathogens and their mechanisms of action. | |||
| Types of nanoparticles | Targeted bacteria | Mechanisms of antibacterial actions | Reference |
| AgNPs | Escherichia coli |
|
[19] |
| Enterococcus faecalis (VRE), Staphylococcus epidermidis (MRSE), Pseudomonas aeruginosa, Staphylococcus aureus |
[13,46] | ||
| Staphylococcus aureus, Pseudomonas aeruginosa, K. pneumoniae, Bacillus cereus Salmonellatyphimurium |
[13,47] | ||
| Escherichia coli, Staphylococcus aureus (MRSA), Streptococcus Pneumoniae | [13,48] | ||
| AuNPs | Escherichia coli, Klebsiella pneumonia |
|
[19,49] |
| Streptococcus Bovis, Staphylococcus epidermidis, Enterococcus Aerogenes |
[19,50] | ||
| ZnONPs | Escherichia coli |
|
[19] |
| Staphylococcus aureus, Pseudomonas aeruginosa |
[30] | ||
| CuNPs | Escherichia coli |
|
[13,19] |
| Staphylococcus aureus, Staphylococcusepidermidis |
[40] | ||
| P.aeruginosa | [13,19] | ||
| CTiO2NPs | S. aureus |
|
[51] |
Nanoantibiotics
Owing to the failure of existing antibiotics to effectively combat bacterial infections, the hunt for unconventional biocides has intensified in recent years, and metallic nanoparticles provide a special potential means of combating ARB [35]. NPs offer a prospective approach to managing infectious diseases caused by MDR bacteria, according to [15]. In this regard, NPs have demonstrated medicinal potential in the future due to their distinct physical and chemical properties [13]. Metal oxide NPs such as copper oxide (CuO), titanium dioxide (CTiO2), and zinc oxide (ZnO) have been shown to be effective against pathogens such as methicillin-resistant Staphylococcus aureus and Escherichia coli.
Silver (Ag) NPs
AgNPs are increasingly used in medicine due to their wide range of antimicrobial activities [36]. The use of AgNPs to treat infectious diseases has shown to be effective against MDR pathogens [1,6]. Since AgNPs are smaller and have a higher surface area to volume ratio, they can interfere with cell membranes and influence bacterial structure and metabolism [19]. AgNP has biocidal activity against a wide variety of gram-positive and gram-negative bacteria [37]. Because of their unique physicochemical properties, AgNPs can inhibit the growth of many bacteria and fungi. They have a high surface area-to-mass ratio, which helps them engage with bacteria physically. AgNPs can have a direct effect on bacterial cell membranes, affecting respiration, metabolism, and proliferation. They can cause the formation of reactive oxygen species (ROS), which enhances their antibacterial effects. AgNPs have recently been discovered to disrupt quorum sensing in bacterial biofilms. Pseudomonas aeruginosa biofilm development was inhibited by myco-fabricated AgNPs (mfAg NPs) [25].
The antibacterial nature of AgNPs is mainly based on the following mechanisms: (a) oxidative stress through the catalysis of reactive ROS formation, (b) interference of DNA replication, and (c) release of Ag ions which bind to electron donor groups in molecules containing sulfur, oxygen, or nitrogen [1,19,37]. In particular, AgNPs can introduce ROS and cell membrane damage to bacteria [19]. The entry of AgNPs induces ROS that will inhibit ATP production and DNA replication. Singh, et al. and Dakal, et al. found that AgNPs have also proven to be effective against biofilms of Staphylococcus aureus [5,38]. Furthermore, Lu, et al. found that both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes via inducing ROS overproduction, triggering a response, and increasing cell membrane permeability [1].
Copper Oxide (CuO) NPs
Copper-containing NPs have been shown to be effective against bacteria, prevent the development of MDR biofilms, and have the ability to act as antimicrobial coating agents. Cu2ONPs cut a hole in the cell membrane, allowing unwanted contaminants to flood through the cell more easily [19]. Copper oxide NPs produce reactive oxygen species (ROS), which can contribute to DNA degradation due to the release of metallic ions. Su et al. investigated the effect on the expression of intracellular proteins and when CuONPs enter bacteria, metabolic functions are affected, such as active transport, electron transfer, and nitrogen metabolism [39]. In addition, Kruk, et al. found that copper NPs accomplished inhibiting the growth of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus [40]. The study conducted by, Zhang, et al. examined and found that Cu2O showed a considerably synergistic antibacterial effect combined with aminoglycoside antibiotics against Escherichia coli, and the inhibition zone area increased by 59.0% when Cu2O was combined with neomycin [19].
Zinc Oxide (ZnO) NPs
ZnO NPs have antibacterial properties against a wide range of bacteria and fungus. ZnO NPs, like Ag NPs, promote the formation of reactive oxygen species (ROS), which leads to cell death. They can also disrupt the integrity of the bacterial cell membrane, resulting in leakage and bacterial death. At a concentration of only 12 g/mL, ZnO NPs can prevent Streptococcus pneumoniae, a major cause of pneumonia, from forming biofilms. ZnO NPs could disrupt oral biofilms through a variety of mechanisms, including (1) disrupting EPS and causing single bacteria to be separated from the biofilm, (2) triggering the production of reactive oxygen species (ROS), (3) binding to and inhibiting the normal functions of DNA and enzymes [25,41]. ZnONPs can inhibit biofilm formation by Streptococcus pneumonia, a common cause of pneumonia, and reduce biofilm formation by uropathogenic Escherichia coli strains. Also, Agarwal, et al. concluded in their experiment that zinc oxide nanoparticles also happen to block or inhibit the zinc ion efflux pump which aids in increasing local zinc ion concentration in a vulnerable situation [30]. Among metal and metal oxide nanoparticles, silver exhibits remarkable antimicrobial activity and is one of the most extensively studied nanomaterials for their antimicrobial activity [27]. Moreover, antimicrobial activities of silver nanoparticles against pathogens causing global concerns such as TB and cholera are also reported.
Titanium Dioxide (CTiO2) NPs
CTiO2NPs are widely utilized in food items, such as sweets and chewing gums with the E171 designation [27]. When exposed to UV light, CTiO2 acts as a photocatalyst, assisting in the production of powerful oxidizing agents. They can break down Listeria monocytogenes biofilms, prevalent food-borne bacteria that cause health problems during food preparation. CTiO2NPs, on the other hand, can limit the development of bacteria and minimize the formation of biofilms [25]. Also, CTiO2NPs have antibacterial action against Enterococcus faecium, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus aureus among pathogens. This is because CTiO2NPs may have antibacterial action against bacteria by causing DNA damage following internalization, inhibiting respiration, peroxiding membrane phospholipids, and generating reactive oxygen species (ROS) [27]. Antimicrobial properties of CTiO2NPs are primarily related to the formation of reactive oxygen species (ROS), notably –OH free radicals [42]. The mechanism suggested for CTiO2 NPs is oxidative stress induced by ROS. As a consequence, ROS causes DNA damage at specific sites [33].
Gold (Au) NPs
The antibacterial function of AuNPs is linked to the inhibition of ATPase production, which leads to a decline in cell metabolism, as well as the inhibition of the ribosome binding subunit to tRNA [43]. For this reason, Shamaila, et al. hypothesized that AuNPs could disrupt the bacterial respiratory chain by destroying nicotinamide [44]. When the surfaces of Au NPs are altered, their antibacterial activity may be reduced [13]. Through the dissipation of bacterial membrane potential and the increase of adenosine triphosphate (ATP) levels, Au-Pt bimetallic NPs show antibacterial action against multidrug-resistant Escherichia coli [15].
Iron Oxide (FeO) NPs
Iron oxide (FeONPs) is well-known in the biomedical industry for its biocompatibility and magnetic properties. Reduced iron and FeO NPs have recently been discovered to have antibacterial properties that destroy bacteria cells by disrupting the bacterial membrane and causing oxidative stress within the cell [42]. In connection, Taylor, et al. discovered that iron oxide nanoparticles are effective at inactivating Staphylococcus and antibiotic-resistant biofilms [45].
Synergetic effects of NPs antibiotics
Antimicrobial pathways that combine NPs and antibiotics have diverse antimicrobial mechanisms to counteract bacteria’s antibiotic resistance mechanisms. Similarly, Zhang, et al. found that AgNPs conjugated with gentamycin reduce the antibiotic equivalents’ minimum inhibitory concentrations against Escherichia coli NPs and have shown synergy which can help to reduce global warming [19]. Antimicrobial potency can be recovered using a mixture of NPs and clinically available antibiotics. Combining polymyxin B, ciprofloxacin, ampicillin, vancomycin, or erythromycin with Ag, Au, or ZnONPs may help reverse antimicrobial resistance in MDROs such as antibiotic-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, and vancomycin-resistant Enterococcus (VRE); and methicillin-resistant S. aureus (MRSA) [13].
Dose optimization and toxicity nature of nanoparticles
For medical translation, determining the optimum dosage is critical for clinical goals and minimizing toxicity. The concentrations of nanomaterials that cause cell damage in vitro are currently unrealistically high and impractical to apply to humans, so results from animal experiments may not be specifically applicable to humans [27,52]. While the precise cause of toxic complications is unknown, it has been found that the bigger the NP, the higher the risk of negative health consequences.
Nanoparticle delivery systems improve drug solubility, provide immune evasion by stealth, modulate drug release characteristics, target drug molecules to specific sites, and deliver several drugs at once [53]. Several forms of drug delivery mechanisms have been investigated over the last five decades to encapsulate medications and other biomolecules in general, comprising liposomes, micelles, and dendrimers [54]. Targeted NP-based drug delivery may reduce antibiotic systemic toxicity, decrease drug uptake, increased efflux, and destroy biofilm formation [13]. Chitosan/CTiO2/Ag nanocomposites showed antibacterial activity by generating ROS, releasing lactate dehydrogenase, and inhibiting bacterial adhesion. Salmonella sp. was resistant to chitosan/Ag nanocomposites [55]. Antibacterial behavior of chitosan doped with Ag+ was observed against Staphylococcus aureus and Pseudomonas aeruginosa [56].
For many years, antibiotics have been saving an enormous number of lives from many infectious diseases caused by disease-causing bacteria, and now antibiotic resistance is increasing at an alarming rate and is widely recognized as a global issue that requires urgent attention. The indiscriminate and inappropriate application of antibiotics in different communities and environments is definitely in charge of alarming the health crisis of antibiotic resistance. For this, there are selective pressures, which lead only the fitting genotype to thrive, which results in the emergence of MDR strains by genetic exchange through different mechanisms and emphasizes the need for alternative therapeutic options. Timely, the emergence of resistant and more virulent strains of bacteria has outpaced the development of new antibiotics over the last few years, and of many different strategies to overcome this problem, NPs both as nanoantibiotics and vehicles seem to hold the highest promise. For this, nanotechnology is an urgent need for a novel therapeutic agent and which is getting used increasingly as both antibacterial and veχtor for adequate therapeutic transport of drugs to specific sites of emerging and reemerging MDR pathogens.
Nanoparticles are engineered with unique properties, and their effect is determined by their small size and large surface area, by which they destroy the membrane and pass through the microbe’s body. For this reason, substantial efforts have been made to improve nanoparticle-based techniques with the primary aim of increasing therapeutic effectiveness while reducing the production of drug resistance. These techniques have shown positive results for local antimicrobial treatment by encouraging antibiotic localization to the pathogen and modulating drug-pathogen interactions.
Within a depth review of the literature, it is recognized that studies have explored the treatment of ARB by nanoparticles, but much has yet to be executed, although there is a resistance mechanism that the pathogens are developing. Although there are achievements in the application of NPs for the treatment of antibiotic-resistant pathogens that have been reported, still there are substantial gaps in research are identified including but not limited to that require to be addressed as future perspectives:
There are considerable numbers of signs of progress in the use of metal oxide-based nanoparticles for the treatment of ARB purposes in vitro that have also been addressed in this review. But there are little or no data on the clinical applications and toxicity of NPs and this is one area of research in the future that could potentially lead to wider applications of nanomedicine in the treatment of multidrug-resistant bacteria.
Currently, there are a few clinical trials that are implementing drugs loaded in nanocarriers, even though there are a higher number of basic research studies that do not go further than in vitro experiments and would be a further area of research.
There could be many opportunities in the shortly soon as nanoantibiotics are becoming more widely and thoroughly researched. Therefore, further investigation is required to set up adequate in vivo models, because more physiological barriers and immunological responses are involved in animals upon exposure to nanoparticles, and most are not yet well understood.
Therefore, investing in nanoantibiotics and nano delivery systems developed for multidrug resistance bacterial treatment might be a way forward in the foreseeable future.
Author’s contributions
Tigabu Bekele Mekonnen: Conceptualization, Writing-original draft, Communication. Getachew Alamnie Achenef: Writing-review and editing, Supervision.
The authors are grateful to the College of Natural and Computational Sciences at Mekdela Amba University for their contributions in the process of developing the review, preparing different workshops, and provision of various services.
Declaration of competing interest
This manuscript entitled “Treatment of Antibiotic-Resistant Bacteria by Nanoparticles: Current Approaches and Prospects” consists of 2 tables and a figure. It has not been published previously; it is not under consideration for publication elsewhere. The manuscript is sent up with the approval of all authors. If accepted, it will not be published elsewhere in the same form, in English, or in any other language, including electronically without the written consent of the copyright holder.
- Lu J, Wang Y, Jin M, Yuan Z, Bond P, Guo J. Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes. Water Res. 2020 Feb 1;169:115229. doi: 10.1016/j.watres.2019.115229. Epub 2019 Oct 25. PMID: 31783256.
- Nguyen BT, Chen QL, He JZ, Hu HW. Microbial regulation of natural antibiotic resistance: Understanding the protist-bacteria interactions for evolution of soil resistome. Sci Total Environ. 2020 Feb 25;705:135882. doi: 10.1016/j.scitotenv.2019.135882. Epub 2019 Dec 2. PMID: 31818598.
- Osman M, Al Mir H, Rafei R, Dabboussi F, Madec JY, Haenni M, Hamze M. Epidemiology of antimicrobial resistance in Lebanese extra-hospital settings: An overview. J Glob Antimicrob Resist. 2019 Jun;17:123-129. doi: 10.1016/j.jgar.2018.11.019. Epub 2018 Dec 12. PMID: 30553113.
- Kabra R, Chauhan N, Kumar A, Ingale P, Singh S. Efflux pumps and antimicrobial resistance: Paradoxical components in systems genomics. Prog Biophys Mol Biol. 2019 Jan;141:15-24. doi: 10.1016/j.pbiomolbio.2018.07.008. Epub 2018 Jul 18. PMID: 30031023; PMCID: PMC7173168.
- Singh R, Smitha MS, Singh SP. The role of nanotechnology in combating multi-drug resistant bacteria. J Nanosci Nanotechnol. 2014 Jul;14(7):4745-56. doi: 10.1166/jnn.2014.9527. PMID: 24757944.
- Möhler JS, Sim W, Blaskovich MAT, Cooper MA, Ziora ZM. Silver bullets: A new lustre on an old antimicrobial agent. Biotechnol Adv. 2018 Sep-Oct;36(5):1391-1411. doi: 10.1016/j.biotechadv.2018.05.004. Epub 2018 May 27. PMID: 29847770.
- Chen H, Chen R, Jing L, Bai X, Teng Y. A metagenomic analysis framework for characterization of antibiotic resistomes in river environment: Application to an urban river in Beijing. Environ Pollut. 2019 Feb;245:398-407. doi: 10.1016/j.envpol.2018.11.024. Epub 2018 Nov 12. PMID: 30453138.
- Dong P, Wang H, Fanga T, Wang Y, Yea Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of environmental ARG. Environment International. 2019; 125: 90-96.
- Wang Y, Jin Y, Chen W, Wang J, Chen H, Sun L, Li X, Ji J, Yu Q, Shen L, Wang B. Construction of nanomaterials with targeting phototherapy properties to inhibit resistant bacteria and biofilm infections. Chemical Engineering Journal. 2019. 358: 74-90.
- Huang W, Zhang Q, Li W, Yuan M, Zhou J, Hua L, Chen Y, Ye C, Ma Y. Development of novel nanoantibiotics using an outer membrane vesicle-based drug efflux mechanism. J Control Release. 2020 Jan 10;317:1-22. doi: 10.1016/j.jconrel.2019.11.017. Epub 2019 Nov 15. PMID: 31738965.
- Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A. 2018 Apr 10;115(15):E3463-E3470. doi: 10.1073/pnas.1717295115. Epub 2018 Mar 26. PMID: 29581252; PMCID: PMC5899442.
- Peterson E, Kaur P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front Microbiol. 2018 Nov 30;9:2928. doi: 10.3389/fmicb.2018.02928. PMID: 30555448; PMCID: PMC6283892.
- Hemeg HA. Nanomaterials for alternative antibacterial therapy. Int J Nanomedicine. 2017 Nov 10;12:8211-8225. doi: 10.2147/IJN.S132163. PMID: 29184409; PMCID: PMC5689025.
- Lima R, Del Fiol FS, Balcão VM. Prospects for the Use of New Technologies to Combat Multidrug-Resistant Bacteria. Front Pharmacol. 2019 Jun 21;10:692. doi: 10.3389/fphar.2019.00692. PMID: 31293420; PMCID: PMC6598392.
- Baptista PV, McCusker MP, Carvalho A, Ferreira DA, Mohan NM, Martins M, Fernandes AR. Nano-Strategies to Fight Multidrug Resistant Bacteria-"A Battle of the Titans". Front Microbiol. 2018 Jul 2;9:1441. doi: 10.3389/fmicb.2018.01441. PMID: 30013539; PMCID: PMC6036605.
- Kelly AS, Rodgers MA, O’Brien CS, Donnelly FR, Gilmore FB. Antibiotic delivery strategies to reduce antimicrobial resistance. Trends in Biotechnology. 2019; 1-17.
- Patra JK, Das G, Fraceto LF, Campos EVR, Rodriguez-Torres MDP, Acosta-Torres LS, Diaz-Torres LA, Grillo R, Swamy MK, Sharma S, Habtemariam S, Shin HS. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnology. 2018 Sep 19;16(1):71. doi: 10.1186/s12951-018-0392-8. PMID: 30231877; PMCID: PMC6145203.
- Pazda M, Kumirska J, Stepnowski P, Mulkiewicz E. Antibiotic resistance genes identified in wastewater treatment plant systems - A review. Sci Total Environ. 2019 Dec 20;697:134023. doi: 10.1016/j.scitotenv.2019.134023. Epub 2019 Aug 22. PMID: 31479900.
- Zhang Y, Yuan Y, Chen W, Fan J, Lv H, Wu Q. Integrated nanotechnology of synergism-sterilization and removing-residues for neomycin through nano-Cu2O. Colloids Surf B Biointerfaces. 2019 Nov 1;183:110371. doi: 10.1016/j.colsurfb.2019.110371. Epub 2019 Jul 30. PMID: 31408783.
- Guo M, Ye J, Gao D, Xu N, Yang J. Agrobacterium-mediated horizontal gene transfer: Mechanism, biotechnological application, potential risk and forestalling strategy. Biotechnol Adv. 2019 Jan-Feb;37(1):259-270. doi: 10.1016/j.biotechadv.2018.12.008. Epub 2018 Dec 21. PMID: 30579929.
- Almakki A, Jumas-Bilak E, Marchandin H, Licznar-Fajardo P. Antibiotic resistance in urban runoff. Sci Total Environ. 2019 Jun 1;667:64-76. doi: 10.1016/j.scitotenv.2019.02.183. Epub 2019 Feb 13. PMID: 30826682.
- Molechan C, Amoako DG, Abia ALK, Somboro AM, Bester LA, Essack SY. Molecular epidemiology of antibiotic-resistant Enterococcus spp. from the farm-to-fork continuum in intensive poultry production in KwaZulu-Natal, South Africa. Sci Total Environ. 2019 Nov 20;692:868-878. doi: 10.1016/j.scitotenv.2019.07.324. Epub 2019 Jul 21. PMID: 31539992.
- Ogawara H. Penicillin-binding proteins in Actinobacteria. J Antibiot (Tokyo). 2015 Apr;68(4):223-45. doi: 10.1038/ja.2014.148. Epub 2014 Oct 29. PMID: 25351947.
- Verni M, Minisci A, Convertino S, Nionelli L, Rizzello CG. Wasted Bread as Substrate for the Cultivation of Starters for the Food Industry. Front Microbiol. 2020 Feb 28;11:293. doi: 10.3389/fmicb.2020.00293. PMID: 32184770; PMCID: PMC7058793.
- Cao Y, Naseri M, He Y, Xu C, Walsh LJ, Ziora ZM. Non-antibiotic antimicrobial agents to combat biofilm-forming bacteria. J Glob Antimicrob Resist. 2020 Jun;21:445-451. doi: 10.1016/j.jgar.2019.11.012. Epub 2019 Dec 10. PMID: 31830536.
- Reen FJ, Gutiérrez-Barranquero JA, Parages ML, O Gara F. Coumarin: a novel player in microbial quorum sensing and biofilm formation inhibition. Appl Microbiol Biotechnol. 2018 Mar;102(5):2063-2073. doi: 10.1007/s00253-018-8787-x. Epub 2018 Feb 1. PMID: 29392389; PMCID: PMC5814477.
- Khan F, Pham DTN, Oloketuyi SF, Manivasagan P, Oh J, Kim YM. Chitosan and their derivatives: Anti-biofilm drugs against pathogenic bacteria. Colloids and Surfaces B: Biointerfaces. 2020; 136: 103673.
- Karami P, Khaledi A, Mashoof RY, Yaghoobi MH, Karami M, Dastan D, Alikhani MY. The correlation between biofilm formation capability and antibiotic resistance pattern in Pseudomonas aeruginosa. Gene Reports. 2020; 18: 100561.
- Saxena V, Pandey LM. Bimetallic assembly of Fe(III) doped ZnO as an effective nanoantibiotic and its ROS independent antibacterial mechanism. J Trace Elem Med Biol. 2020 Jan;57:126416. doi: 10.1016/j.jtemb.2019.126416. Epub 2019 Oct 11. PMID: 31629630.
- Agarwal H, Menon S, Kumar SV, Rajeshkumar S. Mechanistic study on antibacterial action of zinc oxide nanoparticles synthesized using green route. Chemico-Biological Interactions. 2018; 286: 60-70.
- Jelinkova P, Mazumdara A, Sura VP, Kociova S, Dolezelikova K, Jimenez AMJ, Koudelkova Z, Mishra PK, Smerkova K, Heger K, Vaculovicova K, Moulicka M, Adama V. Nanoparticle-drug conjugates treating bacterial infections. Review article. Journal of Controlled Release. 2019; 307: 166-185.
- Mahamuni-Badiger PP, Patil PM, Badiger MV, Patel PR, Thorat-Gadgil BS, Pandit A, Bohara RA. Biofilm formation to inhibition: Role of zinc oxide-based nanoparticles. Mater Sci Eng C Mater Biol Appl. 2020 Mar;108:110319. doi: 10.1016/j.msec.2019.110319. Epub 2019 Oct 23. PMID: 31923962.
- Khezerlou A, Alizadeh-Sani M, Azizi-Lalabadi M, Ehsani A. Nanoparticles and their antimicrobial properties against pathogens including bacteria, fungi, parasites and viruses. Microb Pathog. 2018 Oct;123:505-526. doi: 10.1016/j.micpath.2018.08.008. Epub 2018 Aug 7. PMID: 30092260.
- Anuj SA, Gajera HP, Hirpara DG, Golakiya BA. Bactericidal assessment of nano-silver on emerging and re-emerging human pathogens. J Trace Elem Med Biol. 2019 Jan;51:219-225. doi: 10.1016/j.jtemb.2018.04.028. Epub 2018 Apr 24. PMID: 29735327; PMCID: PMC7126441.
- Shaikh S, Nazam N, Rizvi SMD, Ahmad K, Baig MH, Lee EJ, Choi I. Mechanistic Insights into the Antimicrobial Actions of Metallic Nanoparticles and Their Implications for Multidrug Resistance. Int J Mol Sci. 2019 May 18;20(10):2468. doi: 10.3390/ijms20102468. PMID: 31109079; PMCID: PMC6566786.
- Cheng G, Dai M, Ahmed S, Hao H, Wang X, Yuan Z. Antimicrobial Drugs in Fighting against Antimicrobial Resistance. Front Microbiol. 2016 Apr 8;7:470. doi: 10.3389/fmicb.2016.00470. PMID: 27092125; PMCID: PMC4824775.
- Peters RJ, Bouwmeester H, Gottardo S, Amenta V, Arena M, Brandhoff P. Nanomaterials for products and application in agriculture, feed and food. Trends in Food Science and Technology. 2016; 54: 155-164.
- Dakal TC, Kumar A, Majumdar RS, Yadav V. Mechanistic Basis of Antimicrobial Actions of Silver Nanoparticles. Front Microbiol. 2016 Nov 16;7:1831. doi: 10.3389/fmicb.2016.01831. PMID: 27899918; PMCID: PMC5110546.
- Su Y, Zheng X, Chen Y, Li M, Liu K. Alteration of intracellular protein expressions as a key mechanism of the deterioration of bacterial denitrification caused by copper oxide nanoparticles. Sci Rep. 2015 Oct 28;5:15824. doi: 10.1038/srep15824. PMID: 26508362; PMCID: PMC4623765.
- Kruk T, Szczepanowicz K, Stefańska J, Socha RP, Warszyński P. Synthesis and antimicrobial activity of monodisperse copper nanoparticles. Colloids Surf B Biointerfaces. 2015 Apr 1;128:17-22. doi: 10.1016/j.colsurfb.2015.02.009. Epub 2015 Feb 14. PMID: 25723345.
- Sarwar S, Chakraborti S, Bera S, Sheikh IA, Hoque KM, Chakrabarti P. The antimicrobial activity of ZnO nanoparticles against Vibrio cholerae: Variation in response depends on biotype. Nanomedicine. 2016 Aug;12(6):1499-509. doi: 10.1016/j.nano.2016.02.006. Epub 2016 Mar 10. PMID: 26970029.
- Baranwal A, Srivastava A, Kumar P, Bajpai VK, Maurya PK, Chandra P. Prospects of Nanostructure Materials and Their Composites as Antimicrobial Agents. Front Microbiol. 2018 Mar 9;9:422. doi: 10.3389/fmicb.2018.00422. PMID: 29593676; PMCID: PMC5855923.
- Cui Y, Zhao Y, Tian Y, Zhang W, Lü X, Jiang X. The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials. 2012 Mar;33(7):2327-33. doi: 10.1016/j.biomaterials.2011.11.057. Epub 2011 Dec 17. PMID: 22182745.
- Shamaila S, Zafar N, Riaz S, Sharif R, Nazir J, Naseem S. Gold Nanoparticles: An Efficient Antimicrobial Agent against Enteric Bacterial Human Pathogen. Nanomaterials (Basel). 2016 Apr 14;6(4):71. doi: 10.3390/nano6040071. PMID: 28335198; PMCID: PMC5302575.
- Taylor EN, Kummer KM, Durmus NG, Leuba K, Tarquinio KM, Webster TJ. Superparamagnetic iron oxide nanoparticles (SPION) for the treatment of antibiotic-resistant biofilms. Small. 2012 Oct 8;8(19):3016-27. doi: 10.1002/smll.201200575. Epub 2012 Jul 6. PMID: 22777831.
- Esmaeillou M, Zarrini G, Ahangarzadeh Rezaee M, Shahbazi Mojarrad J, Bahadori A. Vancomycin Capped with Silver Nanoparticles as an Antibacterial Agent against Multi-Drug Resistance Bacteria. Adv Pharm Bull. 2017 Sep;7(3):479-483. doi: 10.15171/apb.2017.058. Epub 2017 Sep 25. PMID: 29071232; PMCID: PMC5651071.
- Otari SV, Patil RM, Waghmare SR, Ghosh SJ, Pawar SH. A novel microbial synthesis of catalytically active Ag-alginate biohydrogel and its antimicrobial activity. Dalton Trans. 2013 Jul 21;42(27):9966-75. doi: 10.1039/c3dt51093j. Epub 2013 May 23. PMID: 23698554.
- Thapa R, Bhagat C, Shrestha P, Awal S, Dudhagara P. Enzyme-mediated formulation of stable elliptical silver nanoparticles tested against clinical pathogens and MDR bacteria and development of antimicrobial surgical thread. Ann Clin Microbiol Antimicrob. 2017 May 16;16(1):39. doi: 10.1186/s12941-017-0216-y. PMID: 28511708; PMCID: PMC5434635.
- Shaikh S, Rizvi SMD, Shakil S, Hussain T, Alshammari TM, Ahmad W, Tabrez S, Al-Qahtani MH, Abuzenadah AM. Synthesis and Characterization of Cefotaxime Conjugated Gold Nanoparticles and Their Use to Target Drug-Resistant CTX-M-Producing Bacterial Pathogens. J Cell Biochem. 2017 Sep;118(9):2802-2808. doi: 10.1002/jcb.25929. Epub 2017 Apr 27. PMID: 28181300.
- Payne JN, Waghwani HK, Connor MG, Hamilton W, Tockstein S, Moolani H, Chavda F, Badwaik V, Lawrenz MB, Dakshinamurthy R. Novel Synthesis of Kanamycin Conjugated Gold Nanoparticles with Potent Antibacterial Activity. Front Microbiol. 2016 May 2;7:607. doi: 10.3389/fmicb.2016.00607. PMID: 27330535; PMCID: PMC4908860.
- Roy A, Parveen AR, Koppalkar A, Prasad MVNA. Effect of nano - titanium dioxide with different antibiotics against methicillin-resistant Staphylococcus aureus. Journal of Biomaterials and Nanobiotechnology. 2010; 1: 37-41.
- Hua S, de Matos MBC, Metselaar JM, Storm G. Current Trends and Challenges in the Clinical Translation of Nanoparticulate Nanomedicines: Pathways for Translational Development and Commercialization. Front Pharmacol. 2018 Jul 17;9:790. doi: 10.3389/fphar.2018.00790. PMID: 30065653; PMCID: PMC6056679.
- Gao W, Chen Y, Zhang Y, Zhang Q, Zhang L. Nanoparticle-based local antimicrobial drug delivery. Adv Drug Deliv Rev. 2018 Mar 1;127:46-57. doi: 10.1016/j.addr.2017.09.015. Epub 2017 Sep 20. PMID: 28939377; PMCID: PMC5860926.
- Wang DY, van der Mei HC, Ren Y, Busscher HJ, Shi L. Lipid-Based Antimicrobial Delivery-Systems for the Treatment of Bacterial Infections. Front Chem. 2020 Jan 10;7:872. doi: 10.3389/fchem.2019.00872. PMID: 31998680; PMCID: PMC6965326.
- Arjunan N, Kumari HL, Singaravelu CM, Kandasamy R, Kandasamy J. Physicochemical investigations of biogenic chitosan-silver nanocomposite as antimicrobial and anticancer agent. Int J Biol Macromol. 2016 Nov;92:77-87. doi: 10.1016/j.ijbiomac.2016.07.003. Epub 2016 Jul 2. PMID: 27381584.
- El-Nahrawy AM, Ali AI, Abou Hammad AB, Youssef AM. Influences of Ag-NPs doping chitosan/calcium silicate nanocomposites for optical and antibacterial activity. Int J Biol Macromol. 2016 Dec;93(Pt A):267-275. doi: 10.1016/j.ijbiomac.2016.08.045. Epub 2016 Aug 16. PMID: 27543348.